(1) Updated the installation method for DeepST.
(2) Fixed some bugs.
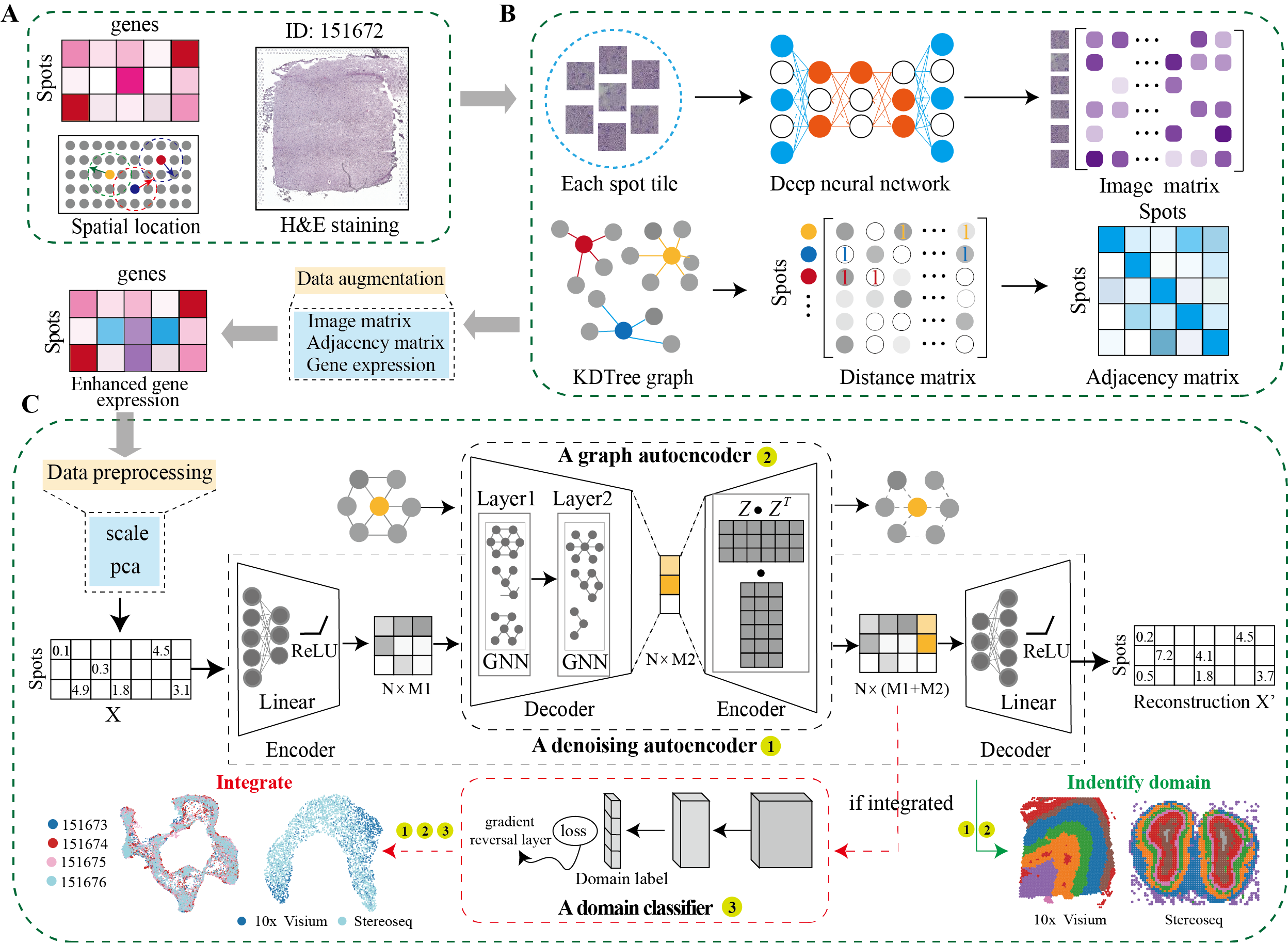
DeepST first uses H&E staining to extract tissue morphology information through a pre-trained deep learning model, and normalizes each spot’s gene expression according to the similarity of adjacent spots. DeepST further learns a spatial adjacency matrix on spatial location for the construction of graph convolutional network. DeepST uses a graph neural network autoencoder and a denoising autoencoder to jointly generate a latent representation of augmented ST data, while domain adversarial neural networks (DAN) are used to integrate ST data from multi-batches or different technologies. The output of DeepST can be applied to identify spatial domains, batch effect correction and downstream analysis.
To install DeepST, make sure you have PyTorch and PyG installed. For more details on dependencies, refer to the environment.yml file.
conda create -n deepst-env python=3.9
Activate the environment and install PyTorch and PyG. Adjust the installation commands based on your CUDA version or choose the CPU version if necessary.
- General Installation Command
conda activate deepst-env
pip install torch==2.1.0 torchvision==0.16.0 torchaudio==2.1.0 --index-url https://download.pytorch.org/whl/cu118
pip install pyg_lib==0.3.1+pt21cu118 torch_scatter torch_sparse torch_cluster torch_spline_conv -f https://data.pyg.org/whl/torch-2.1.0+cu118.html
pip install torch_geometric==2.3.1
- Tips for selecting the correct CUDA version
- Run the following command to verify CUDA version:
nvcc --version- Alternatively, use:
nvidia-smi - Modify installation commands based on CUDA
- For CUDA 12.1
pip install torch==2.1.0 torchvision==0.16.0 torchaudio==2.1.0 --index-url https://download.pytorch.org/whl/cu121 pip install pyg_lib==0.3.1+pt21cu121 torch_scatter torch_sparse torch_cluster torch_spline_conv -f https://data.pyg.org/whl/torch-2.1.0+cu121.html pip install torch_geometric==2.3.1 - For CPU-only
pip install torch==2.1.0 torchvision==0.16.0 torchaudio==2.1.0 --index-url https://download.pytorch.org/whl/cpu pip install pyg_lib==0.3.1+pt21cpu torch_scatter torch_sparse torch_cluster torch_spline_conv -f https://data.pyg.org/whl/torch-2.1.0+cpu.html pip install torch_geometric==2.3.1
- For CUDA 12.1
pip install deepstkit
import deepstkit as dt
import os
import matplotlib.pyplot as plt
import scanpy as sc
import deepstkit as dt
# ========== Configuration ==========
SEED = 0 # Random seed for reproducibility
DATA_DIR = "../data/DLPFC" # Directory containing spatial data
SAMPLE_ID = "151673" # Sample identifier to analyze
RESULTS_DIR = "../Results" # Directory to save outputs
N_DOMAINS = 7 # Expected number of spatial domains
# ========== Initialize Analysis ==========
# Set random seed and initialize DeepST
dt.utils_func.seed_torch(seed=SEED)
# Create DeepST instance with analysis parameters
deepst = dt.main.run(
save_path=RESULTS_DIR,
task="Identify_Domain", # Spatial domain identification
pre_epochs=500, # Pretraining iterations
epochs=500, # Main training iterations
use_gpu=True # Accelerate with GPU if available
)
# ========== Data Loading & Preprocessing ==========
# (Optional) Load spatial transcriptomics data (Visium platform)
# e.g. adata = anndata.read_h5ad("*.h5ad"), this data including .obsm['spatial']
adata = deepst._get_adata(
platform="Visium",
data_path=DATA_DIR,
data_name=SAMPLE_ID
)
# Optional: Incorporate H&E image features (skip if not available)
# adata = deepst._get_image_crop(adata, data_name=SAMPLE_ID)
# ========== Feature Engineering ==========
# Data augmentation (skip morphological if no H&E)
adata = deepst._get_augment(
adata,
spatial_type="BallTree",
use_morphological = False # Set True if using H&E features
)
# Construct spatial neighborhood graph
graph_dict = deepst._get_graph(
adata.obsm["spatial"],
distType="KDTree" # Spatial relationship modeling
)
# Dimensionality reduction
data = deepst._data_process(
adata,
pca_n_comps=200 # Reduce to 200 principal components
)
# ========== Model Training ==========
# Train DeepST model and obtain embeddings
deepst_embed = deepst._fit(
data=data,
graph_dict=graph_dict
)
adata.obsm["DeepST_embed"] = deepst_embed
# ========== Spatial Domain Detection ==========
# Cluster spots into spatial domains
adata = deepst._get_cluster_data(
adata,
n_domains=N_DOMAINS, # Expected number of domains
priori=True # Use prior knowledge if available
)
# ========== Visualization & Output ==========
# Plot spatial domains
sc.pl.spatial(
adata,
color=["DeepST_refine_domain"], # Color by domain
frameon=False,
spot_size=150,
title=f"Spatial Domains - {SAMPLE_ID}"
)
# Save results
output_file = os.path.join(RESULTS_DIR, f"{SAMPLE_ID}_domains.pdf")
plt.savefig(output_file, bbox_inches="tight", dpi=300)
print(f"Analysis complete! Results saved to {output_file}")import os
import matplotlib.pyplot as plt
import scanpy as sc
import deepstkit as dt
# ========== Configuration ==========
SEED = 0
DATA_DIR = "../data/DLPFC"
SAMPLE_IDS = ['151673', '151674','151675', '151676']
RESULTS_DIR = "../Results"
N_DOMAINS = 7
INTEGRATION_NAME = "_".join(SAMPLE_IDS)
# ========== Initialize Analysis ==========
# Set random seed and initialize DeepST
dt.utils_func.seed_torch(seed=SEED)
# ========== Initialize DeepST Integration ==========
integration_model = dt.main.run(
save_path=RESULTS_DIR,
task="Integration", # Multi-sample integration task
pre_epochs=500,
epochs=500,
use_gpu=True
)
# ========== Multi-Sample Processing ==========
processed_data = []
spatial_graphs = []
for sample_id in SAMPLE_IDS:
# Load and preprocess each sample
adata = integration_model._get_adata(
platform="Visium",
data_path=DATA_DIR,
data_name=sample_id
)
# Incorporate H&E image features (Optional)
# adata = integration_model._get_image_crop(adata, data_name=sample_id)
# Feature augmentation
adata = integration_model._get_augment(
adata,
spatial_type="BallTree",
use_morphological=False, # Use prior knowledge if available
)
# Construct spatial neighborhood graph
graph = integration_model._get_graph(
adata.obsm["spatial"],
distType="KDTree"
)
processed_data.append(adata)
spatial_graphs.append(graph)
# ========== Dataset Integration ==========
# Combine multiple samples into integrated dataset
combined_adata, combined_graph = integration_model._get_multiple_adata(
adata_list=processed_data,
data_name_list=SAMPLE_IDS,
graph_list=spatial_graphs
)
# Dimensionality reduction
integrated_data = integration_model._data_process(
combined_adata,
pca_n_comps=200
)
# ========== Integrated Model Training ==========
# Train with domain adversarial learning
embeddings = integration_model._fit(
data=integrated_data,
graph_dict=combined_graph,
domains=combined_adata.obs["batch"].values, # For batch correction
n_domains=len(SAMPLE_IDS) ) # Number of batches
combined_adata.obsm["DeepST_embed"] = embeddings
# ========== Spatial Domain Detection ==========
combined_adata = integration_model._get_cluster_data(
combined_adata,
n_domains=N_DOMAINS,
priori=True, # Use biological priors if available
batch_key="batch_name",
)
# ========== Visualization ==========
# UMAP of integrated data
sc.pp.neighbors(combined_adata, use_rep='DeepST_embed')
sc.tl.umap(combined_adata)
# Save combined UMAP plot
umap_plot = sc.pl.umap(
combined_adata,
color=["DeepST_refine_domain", "batch_name"],
title=f"Integrated UMAP - Samples {INTEGRATION_NAME}",
return_fig=True
)
umap_plot.savefig(
os.path.join(RESULTS_DIR, f"{INTEGRATION_NAME}_integrated_umap.pdf"),
bbox_inches='tight',
dpi=300
)
# Save individual spatial domain plots
for sample_id in SAMPLE_IDS:
sample_data = combined_adata[combined_adata.obs["batch_name"]==sample_id]
spatial_plot = sc.pl.spatial(
sample_data,
color='DeepST_refine_domain',
title=f"Spatial Domains - {sample_id}",
frameon=False,
spot_size=150,
return_fig=True
)
spatial_plot.savefig(
os.path.join(RESULTS_DIR, f"{sample_id}_domains.pdf"),
bbox_inches='tight',
dpi=300
)
print(f"Integration complete! Results saved to {RESULTS_DIR}")Tools that are compared include:
| Platform | Tissue | SampleID |
|---|---|---|
| 10x Visium | Human dorsolateral pre-frontal cortex (DLPFC) | 151507, 151508, 151509, 151510, 151669, 151670, 151671, 151672, 151673, 151674, 151675, 151676 |
| 10x Visium | Mouse brain section | Coronal, Sagittal-Anterior, Sagittal-Posterior |
| 10x Visium | Human breast cancer | Invasive Ductal Carcinoma breast, Ductal Carcinoma In Situ & Invasive Carcinoma |
| Stereo-Seq | Mouse olfactory bulb | Olfactory bulb |
| Slide-seq | Mouse hippocampus | Coronal |
| MERFISH | Mouse brain slice | Hypothalamic preoptic region |
Spatial transcriptomics data of other platforms can be downloaded https://www.spatialomics.org/SpatialDB/
Feel free to submit an issue or contact us at xuchang0214@163.com for problems about the packages.